Single vessel + non-cohesive sediment¶
In this example, you will add sediment morphology to a single vessel ship wakes example. The primary “input.txt” and vessel_00001 files are located in /simple_cases/single_vessel_morphology/.
Model setup
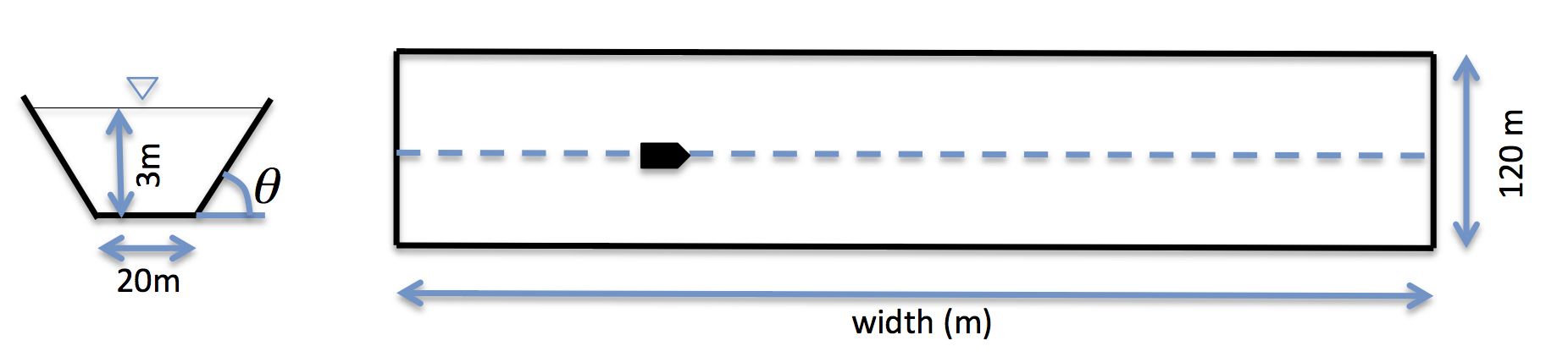
Setup “input.txt”
This example is slightly different from the previous example described in Single vessel + sediment (small domain for training session). Update the following sections:
Set a descriptive title for your simulation:
!-----TITLE----- TITLE = vessel_sediment_morphIf running in parallel, set the number of processors in X and Y:
!-----PARALLEL INFO----- PX = 4 PY = 1Set the bathymetry to the provided depth file:
!-----DEPTH----- DEPTH_TYPE = DATA DEPTH_FILE = ./depth.txtIf a “depth.txt” is not located in the working directory, you will need to run the “mk_depth.f” Fortran script to generate a depth file, which uses the “bathy_param.txt” file as an input. You can do this by typing the following into your terminal:
gfortran mk_depth.f. Refer to Grid and Computational Time for parameter definitions.Send the results to a folder named “output”:
!-----PRINT----- RESULT_FOLDER = output/Set the dimension of the domain to 2400 x 60 (x and y directions, respectively):
!-----DIMENSION----- Mglob = 2400 Nglob = 60Set the computational time, plot time, station printing interval, and screen interval to 2000.0 s, 5.0 s, 50,000.0 s, and 5.0 s, respectively:
!-----TIME----- TOTAL_TIME = 2000.0 PLOT_INTV = 5.0 PLOT_INTV_STATION = 50000.0 SCREEN_INTV = 5.0Keep the single vessel characteristics and sediment morphology parameters as Single vessel + sediment (small domain for training session).
Postprocessing
For postprocessing examples, MATLAB and Python scripts are located in /simple_cases/single_vessel_morphology/postprocessing/. An example model result is shown below:
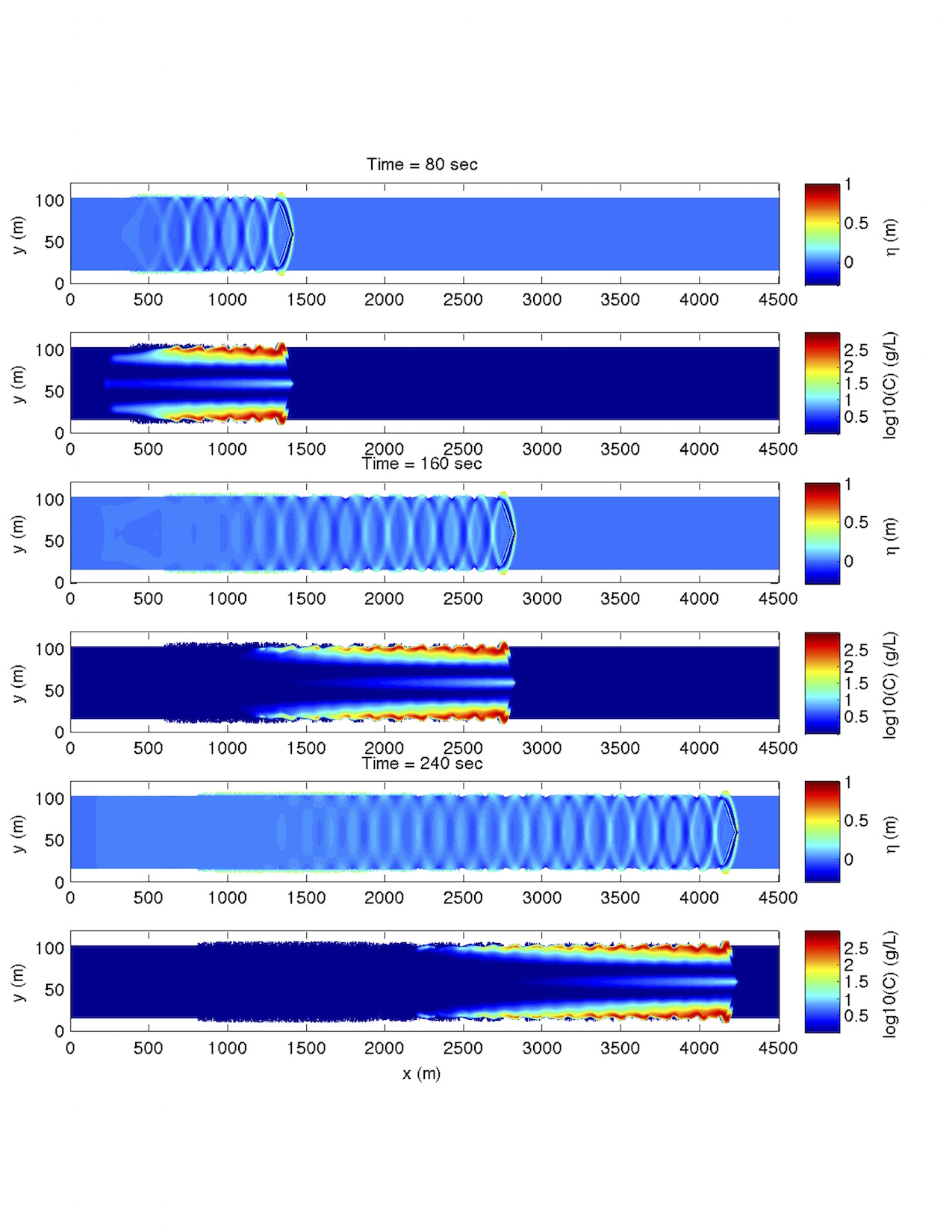
Fig. 12 Shipwakes and sediment concentration at different times.¶
